This server provides a Model Context Protocol (MCP) interface for querying Biomart biological databases, allowing you to:
Discover available Biomart databases (marts) and datasets within them
Explore commonly used or all available attributes and filters for specific datasets
Retrieve biological data using specified attributes and filters
Translate between biological identifiers (e.g., gene symbols to Ensembl IDs) for single or batch operations
Click on "Install Server".
Wait a few minutes for the server to deploy. Once ready, it will show a "Started" state.
In the chat, type
@followed by the MCP server name and your instructions, e.g., "@Biomart MCPtranslate these gene symbols to Ensembl IDs"
That's it! The server will respond to your query, and you can continue using it as needed.
Here is a step-by-step guide with screenshots.
Biomart MCP
A MCP server to interface with Biomart
Model Context Protocol (MCP) is an open protocol that standardizes how applications provide context to LLMs developed by Anthropic. Here we use the MCP python-sdk to create a MCP server that interfaces with Biomart via the pybiomart package.
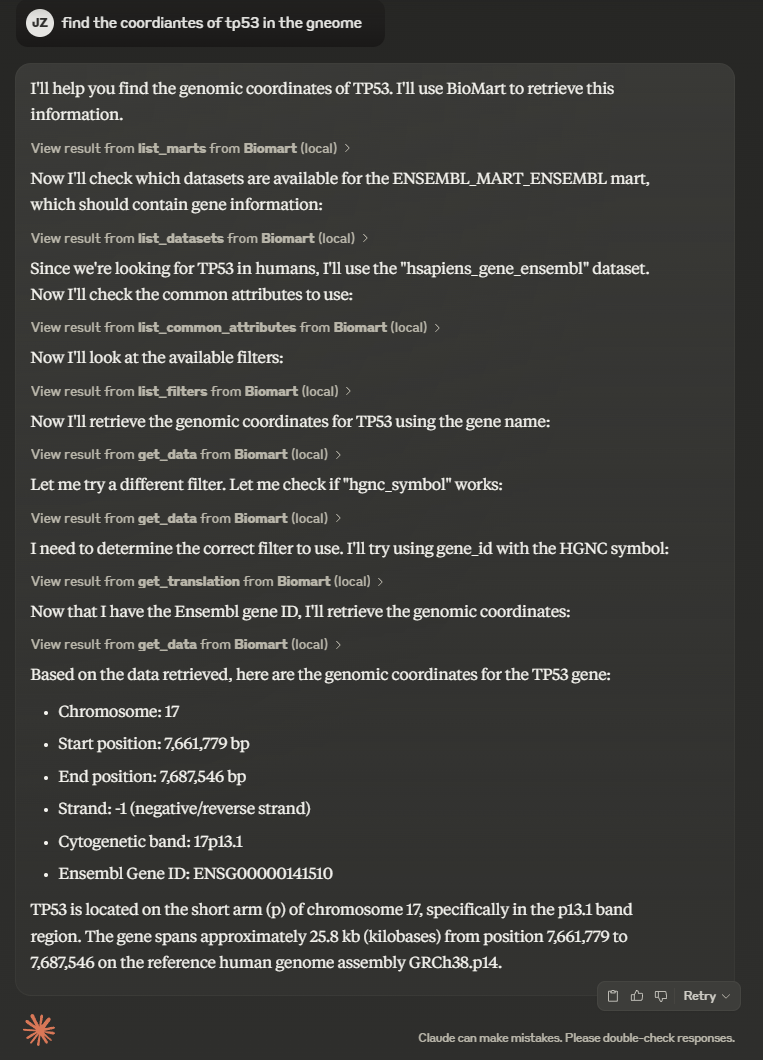
There is a short demo video showing the MCP server in action on Claude Desktop.
Installation
Installing via Smithery
To install Biomart MCP for Claude Desktop automatically via Smithery:
Clone the repository
Claude Desktop
Cursor
Via Cusror's agent mode, other models can take advantage of MCP servers as well, such as those form OpenAI or DeepSeek. Click the cursor setting cogwheel and naviagate to MCP and either add the MCP server to the global config or add it to the a project scope by adding .cursor/mcp.json to the project.
Example .cursor/mcp.json:
Glama
Development
Related MCP server: MCP TapData Server
Features
Biomart-MCP provides several tools to interact with Biomart databases:
Mart and Dataset Discovery: List available marts and datasets to explore the Biomart database structure
Attribute and Filter Exploration: View common or all available attributes and filters for specific datasets
Data Retrieval: Query Biomart with specific attributes and filters to get biological data
ID Translation: Convert between different biological identifiers (e.g., gene symbols to Ensembl IDs)
Contributing
Pull requests are welcome! Some small notes on development:
We are only using
@mcp.tool()here by design, this is to maximize compatibility with clients that support MCP as seen in the docs.We are using
@lru_cacheto cache results of functions that are computationally expensive or make external API calls.We need to be mindful to not blow up the context window of the model, for example you'll see
df.to_csv(index=False).replace("\r", "")in many places. This csv style return is much more token efficient than something likedf.to_string()where the majority of the tokens are whitespace. Also be mindful of the fact that pulling all genes from a chromosome or similar large request will also be too large for the context window.
Potential Future Features
There of course many more features that could be added, some maybe beyond the scope of the name biomart-mcp. Here are some ideas:
Add webscraping for resource sites with
bs4, for example we got the Ensembl gene ID for NOTCH1 then maybe in some cases it would be usful to grap the collatedComments and Description Text from UniProtKBsection from it's page on UCSC$...$